~ナノスケールの動態を‘直接’観る~
サンプルスキャン型高速原子間力顕微鏡 SS-NEX
 |
 |
|
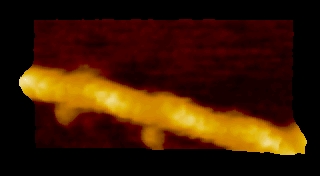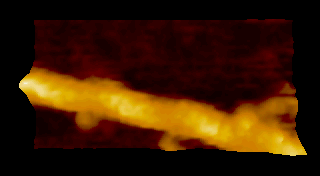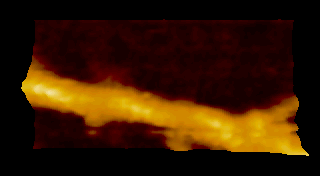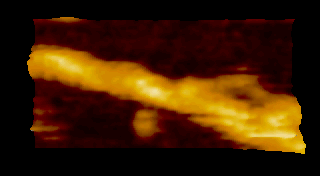
‘歩く’ミオシン V
N. Kodera et al. 2010 |
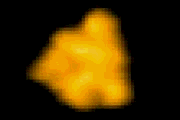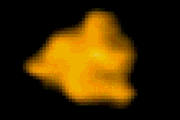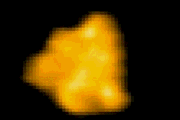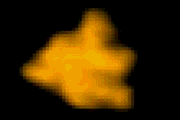
回転軸を取り除いた
F1-ATPase (分子モーター) の‘回転’構造変化 T. Uchihashi et al. 2011 |
高速原子間力顕微鏡(AFM*)は、ナノスケールのサンプルを大気中でも溶液中でも「動画」で可視化できる顕微鏡です。
従来型AFM には、静止画でしか撮れないという重大な欠点がありました。
金沢大学教授・安藤敏夫先生により開発された弊社の 高速AFM NanoExplorer** (NEX) -Ando model- は、
従来型AFM の「走査速度の遅さ」を克服し、リアルタイム動画測定を実現しました。
短時間で画像を取得できるため、サンプルの揺らぎや振動に強く、基板への強固な固定が不要です。
生体分子などのサンプルの反応性を損なうことなく測定できます。
* Atomic Force Microscope (AFM) : 原子間力顕微鏡
** NanoExplorerは、(株) 生体分子計測研究所の登録商標です。
 |
 |
|
抗体IgG
|
HeLa細胞
|
[ このページの先頭に戻る ]
- 独自の共振防止メカニズムにより、
従来型AFMの1000倍以上の速度での高速走査ができます - XYZ三軸の各ピエゾを独立して駆動させることで、
高速走査でも歪みが少ないイメージングを実現しました - 標準型スキャナ、広域型スキャナ、超広域型スキャナを目的に合わせて選択できます
| 標準型スキャナ |
速い走査速度が必要な酵素反応や構造変化の測定に適します
|
|---|---|
| 広域型スキャナ |
水平方向、高さ方向ともに標準型スキャナより広い領域の測定に適します
|
| 超広域型スキャナ |
水平方向、高さ方向ともに広域型スキャナより広い領域の測定に適します
|
注1: スキャナの走査範囲は代表値です
注2: 走査速度は各スキャナ毎に決められた条件下での値であり、最大走査範囲での走査速度を保証するものではありません
- 高共振周波数でありながら低いばね定数を持ったカンチレバーを採用しました
- 生体分子などの柔らかいサンプルに損傷を与えることなく高速走査ができます
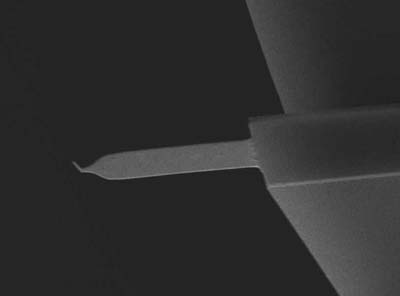
高速AFM用極微小カンチレバー
共振周波数: 大気中 1500 kHz, 溶液中 500 kHz
ばね定数: 0.1 N / m
先端曲率半径: < 10 nm
- 広帯域アナログフィードバック機構に加え、独自のダイナミックPIDを採用しました
- 高速走査下でもサンプル表面に忠実な画像を取得できます
[ このページの先頭に戻る ]
| 走査速度 | 50 ms / frame(20 frames / sec) |
|---|---|
| 最大走査範囲 | X: 0.7 μm, Y: 0.7 μm, Z: 0.4 μm |
| サンプルサイズ | 直径 1.5 mm |
| プローブ検出方式 | 光学検出方式(光てこ式) |
| 走査方式 | サンプルスキャン |
| 測定環境 | 溶液中(測定中に溶液注入可能) / 大気中 |
| 制御方式 | PID/ダイナミックPID制御 |
| 測定モード | AC モード (形状像、誤差像、位相像) |
| その他機能 | スキャナアクティブダンピング、励振効率ドリフト補償 |
注1: 標準型スキャナ使用の場合の仕様です
注2: スキャナの走査範囲は代表値です
注3: 走査速度は各スキャナ毎に決められた条件下での値であり、最大走査範囲での走査速度を保証するものではありません
[ このページの先頭に戻る ]
1. 通常オプション
| 灌流ユニット |
|
|---|---|
| 温度調節ユニット |
|
| 光照射ユニット |
|
2. カンチレバーホルダ
- 「観察溶液を入れる液だめ」と「カンチレバーの固定部」を有したホルダです
- ご要望に合わせて設計・素材(銅、アクリルなど)を変更できます
ご相談ください
| 標準タイプ |
|
|---|---|
| ばねタイプ |
|
| PEEKタイプ |
※形状は標準タイプと同じです |
| 有機溶媒対応 |
|
注: オプションでは、交換可能な上部パーツのみ取り扱っています
[ このページの先頭に戻る ]

